Imperfect pathogen detection from non-invasive skin swabs biases disease inference
Abstract/Summary
1. Conservation managers rely on accurate estimates of disease parameters, such as pathogen prevalence and infection intensity, to assess disease status of a host population. However, these disease metrics may be biased if low-level infection intensities are missed by sampling methods or laboratory diagnostic tests. These false negatives underestimate pathogen prevalence and overestimate mean infection intensity of infected individuals.
2. Our objectives were two-fold. First, we quantified false negative error rates of Batrachochytrium dendrobatidis on non-invasive skin swabs collected from an amphibian community in El Cop?, Panama. We swabbed amphibians twice in sequence, and we used a recently developed hierarchical Bayesian estimator to assess disease status of the population. Second, we developed a novel hierarchical Bayesian model to simultaneously account for imperfect pathogen detection from field sampling and laboratory diagnostic testing. We evaluated the performance of the model using simulations to quantify the magnitude of bias in estimates of pathogen prevalence and infection intensity.
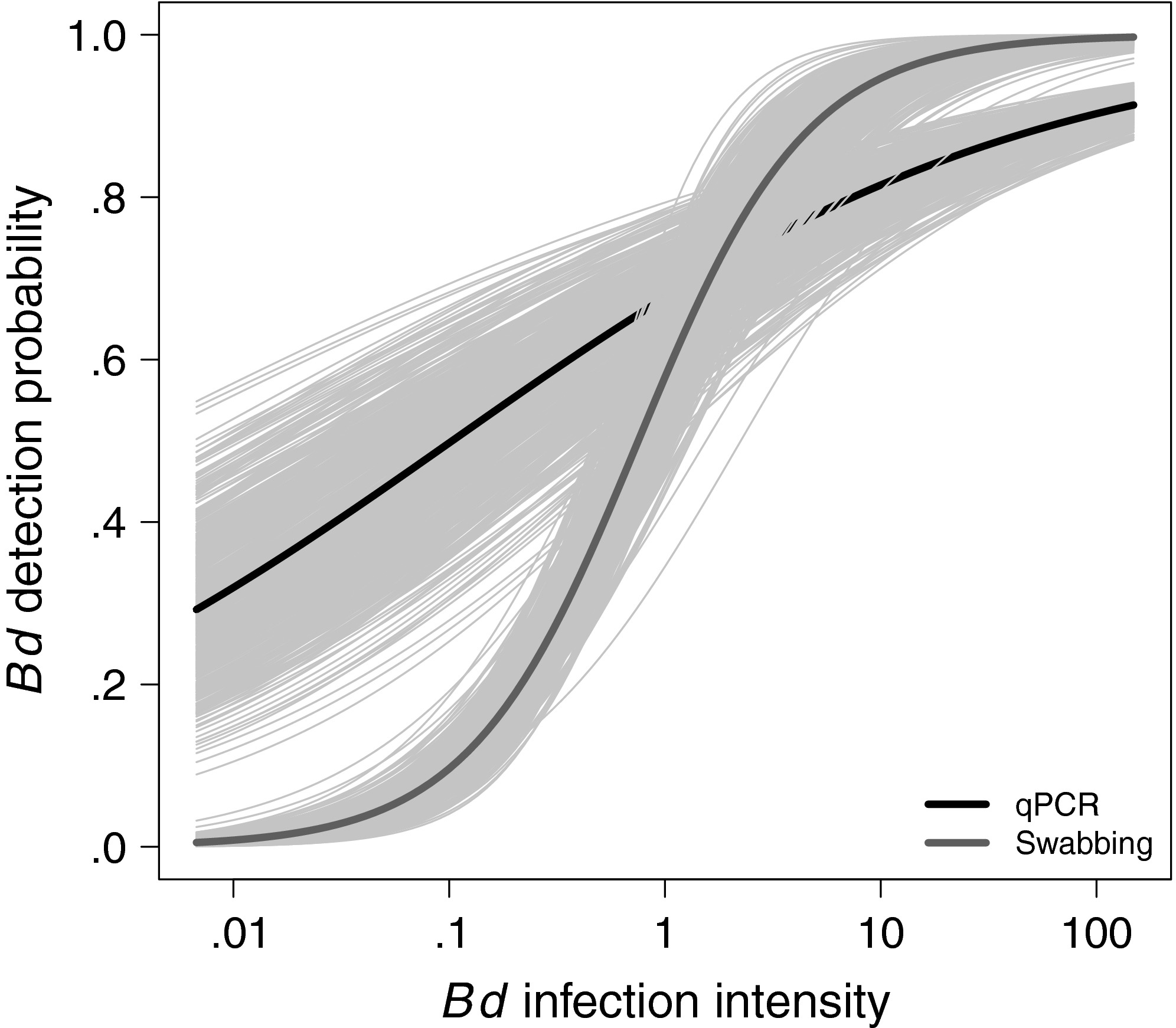
3. We show that Bd detection probability from skin swabs was related to host infection intensity, where Bd infections < 10 zoospores have < 95 % probability of being detected. If imperfect Bd detection was not considered, then Bd prevalence was underestimated by as much as 16%. In the Bd-amphibian system, this indicates a need to correct for imperfect pathogen detection. More generally, our results have implications for study designs in other disease systems, particularly those with similar objectives, biology, and sampling decisions.
4. Uncertainty in pathogen detection is an inherent property of most sampling protocols and diagnostic tests, where the magnitude of bias depends on the study system, type of infection, and false negative error rates. Given that it may be difficult to know this information in advance, we advocate that the most cautious approach is to assume all errors are possible and to accommodate them by adjusting sampling designs. The modeling framework presented here improves the accuracy in estimating pathogen prevalence and infection intensity.
Publication details
| Published Date: | 2017-08-12 |
| Outlet/Publisher: | Methods in Ecology and Evolution |
| Media Format: |
ARMI Organizational Units:
Northeast - BiologyTopics:
Quantitative DevelopmentsKeywords:
detection probabilitydisease
pathogen
sampling design