Estimating occurrence, prevalence, and detection of amphibian pathogens: insights from occupancy models
Abstract/Summary
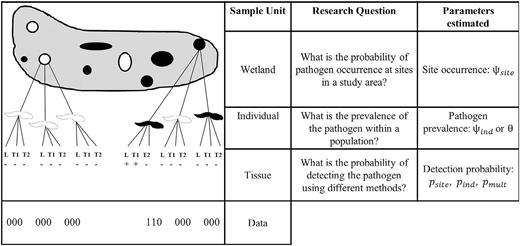
Understanding the distribution of pathogens across landscapes and within host populations is a common aim of wildlife managers. Despite the need for unbiased inferences about these parameters to plan effective management interventions, many researchers fail to account for imperfect pathogen detection and instead report only raw data, which may lead to improper management. We demonstrate analyses of ranavirus detection data in the Patuxent Research Refuge using an occupancy modeling approach, which yields unbiased estimates of pathogen occurrence and prevalence. To improve inference and reporting of pathogen and disease parameters, we describe our approach in-text and provide a written tutorial on how to fit these models using the freely available software Program PRESENCE. In our case study, ranavirus prevalence was underestimated up to 30% if imperfect detection was ignored. After accounting for imperfect detection, estimates of ranavirus prevalence in larval wood frogs (Lithobates sylvaticus) were higher than in larval spotted salamanders (Ambystoma maculatum). In addition, we found that the odds of detecting ranavirus in tail samples were 6.7 times higher than detecting ranavirus in liver samples. We discuss how this information can be used to design ranavirus surveillance studies. Our tutorial provides a clear guide for practitioners and researchers wishing to make unbiased inference in a variety of host-pathogen systems.
Publication details
| Published Date: | 2019 |
| Outlet/Publisher: | Journal of Wildlife Diseases |
| Media Format: |
ARMI Organizational Units:
Northeast - BiologyTopics:
DiseaseQuantitative Developments
Keywords:
detection probabilitydisease
occupancy
Ranavirus